Psychologists nowadays have happily stumbled upon vast reservoirs of ace Python libraries that make bioinformatics and computational biology algorithm-building super-duper easily breezy. These fantabulous libraries are tubular for analyzing biological data sequences, aligning DNA/RNA/protein sequences, calculating mutation rates, drawing phylogenetic trees, motif searching, predicting RNA secondary structure, and more quasi-qualitative analyses. I’m not talking out of my caboose here, these libraries are the cat’s meow according to leading genomics researchers. For that reason, we are here, with a list of the top 10 Python libraries for biology and bioinformatics computational process.
1. BioPython
BioPython is hands down the most bodacious Python library for bioinformatics and computational biology. It’s got a metric buttload of modules for reading/writing different bioinformatics file formats, searching databases, alignment tools, sequence analysis, dealing with ontologies, and BLAST interfacing capabilities. From my personal experience, Biopython makes life a breeze for bioinformaticians. It’s like having a Swiss army knife stuffed with handy dandy tools like parsers, translators, clustering, and machine learning algorithms ready for any kind of bioinformatics problem you wanna throw at it. I think it’s downright awesome.

2. PyCogent
PyCogent is another gnarly Python library for bioinformatics and computational biology applications. This nifty library has got it all – specialized data structures for biological sequences, built-in capabilities for reading/writing files, aligning sequences, inferring phylogenies, estimating evolutionary distances, calculating diversity indices, and a bunch more. PyCogent also integrates nicely with other Python tools like NumPy, SciPy, and Matplotlib. From my perspective, PyCogent takes the cake for sequence alignment and phylogenetics work. Its math methods for DNA, RNA, and protein evolution make tree building and diversity analysis a total piece of cake. I find it to be an invaluable Swiss army knife for molecular evolution research. Rad tool in my opinion.

3. Biskit
Lastly, we have the funkadelic Biskit library. Now this is a gnarly toolbox specialized for structural bioinformatics and molecular modeling. Ya dig? Biskit has all these phat modules for PDB handling, structure analysis, ensemble docking, complex building, MD simulations, and more futuristic stuff. Flexibility analysis, surface patch docking, complex clustering functionality – ya name it and Biskit has some sick tool for it. In my judgment, Biskit is ideal for modeling protein complexes and simulating molecular dynamics. Its integration with other Python libraries like NumPy and PyCogent takes structural bioinformatics to the next level. I think Biskit is thebomb.com for anyone doing structural biology research.
4. Galaxy
Accessible, transparent bioinformatics requires robust data integration, analysis, and visualization frameworks. The Galaxy platform provides these capabilities through a web-based interface. The Galaxy Project offers public servers, cloud options, and Docker images to lower usage barriers. Over 10,000 tools are available for genomics, proteomics, transcriptomics, and more. The Galaxy Python API enables programmatically scripting workflows, modifying configurations, and extending functionality. Galaxy powers reproducible, sharable, and scalable bioinformatics.
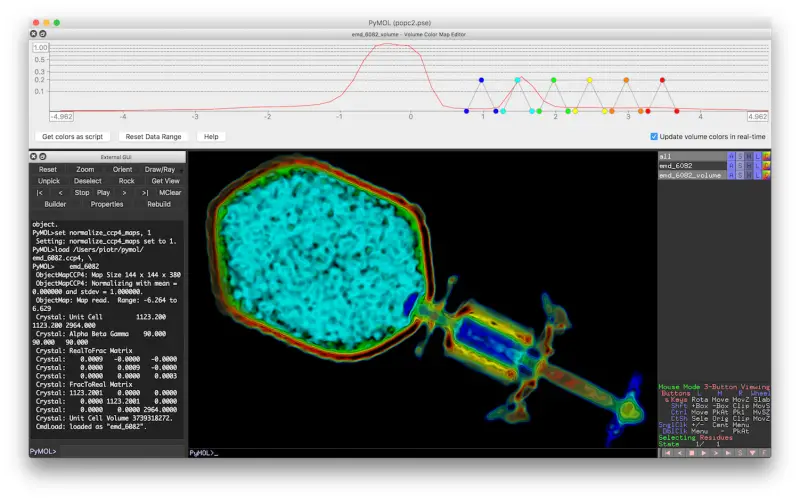
5. PyMOL
PyMOL brings powerful molecular visualization capabilities to Python. This library wraps the popular PyMOL molecular graphics system for scripting automation and extension. PyMOL generates high-quality 3D images and animations of molecular structures and simulations. It offers detailed atomistic rendering with various customization options. Through PyMOL, researchers can flexibly explore macromolecules and simulation trajectories. The object-oriented Python APIs simplify scene setup, molecular styling, movie generation, and more.
6. MDTraj
MDTraj assists rapid analysis of molecular dynamics trajectories with Python. Molecular simulations produce detailed time-series datasets describing atomic interactions. MDTraj handles loading trajectories in common formats like HDF5, netCDF, PDB, TRR, and DCD. It calculates scalar and vector properties for simulation compartments and molecules. Analyses harness the power of NumPy and SciPy for optimal performance. Interactive demos illustrate core capabilities like minimal RMSD clustering. MDTraj streamlines accessing and investigating dynamics.
7. MDAnalysis
MDAnalysis furnishes tools for analyzing molecular dynamics simulations with Python. Like MDTraj, MDAnalysis handles typical trajectory and topology formats. It uses NumPy to enable analyzing time-series data and deriving observables. MDAnalysis provides sophisticated atom selection capabilities for filtering biologically relevant data. Visualization integrates readily with PyMOL, VMD, and Matplotlib. Both rapid scripting and advanced interactive analysis are accessible through its object-oriented design. MDAnalysis combines power, simplicity, and extensibility for MD simulation investigation.

8. Scikit-bio
Scikit-bio provides data structures, algorithms, and educational resources for bioinformatics. This well-designed library focuses on usability, performance, and interoperability. Scikit-bio’s submodules address common tasks like sequence analysis, alignment, tree construction, taxonomy mapping, and molecular evolution simulations. The API follows Scikit-learn’s conventions while functionality resembles Biopython. Scikit-bio is ideal for students and researchers favoring Python for scripting and interactive computing. The API follows Scikit-learn’s style which will be familiar to many Pythonistas today. From my experience, Scikit-bio hits a nice balance between Biopython’s robustness and Pandas’ interactivity.

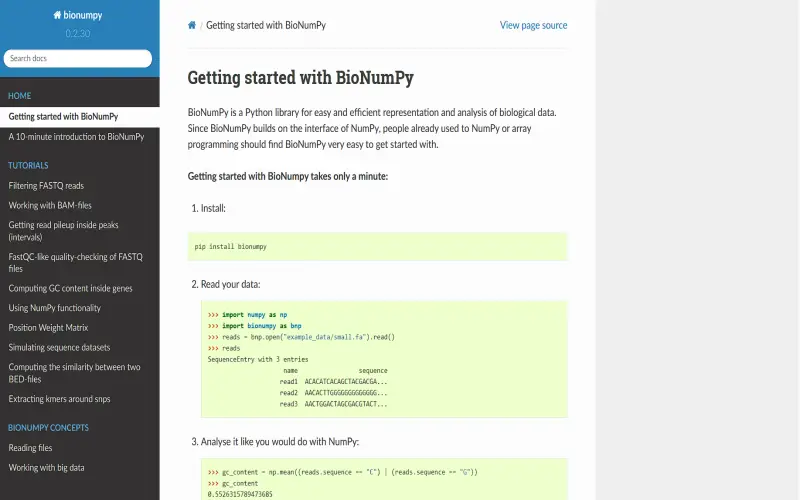
9. BioNumpy
BioNumPy adapts NumPy, Python’s foundational library for scientific computing, for bioinformatics applications. NumPy offers versatile n-dimensional array objects and mathematical functions that underpin many Python scientific libraries. BioNumPy provides domain-specific extensions for biological data like genomic and protein sequences. Users benefit from NumPy’s high-performance thanks to its C and Fortran codebase. BioNumPy unlocks NumPy’s capabilities for bioinformatics while avoiding reinventing wheels. This library essentially adapts NumPy, which provides powerful n-dimensional arrays and math functions, for working with biological data. For example, BioNumPy lets you represent genomic or protein sequences as specialized NumPy arrays. This opens up all of NumPy’s speed and capabilities for bioinformatics tasks without reinventing the wheel.
10. BioPandas
BioPandas enables exploring biological molecules through pandas-like DataFrames and Series. This library facilitates exploratory data analysis approaches popular in modern data science. BioPandas utilizes core pandas data structures to represent and manipulate biological entities like protein structures, genomes, and molecular dynamics trajectories. Users can leverage pandas’ powerful indexing and grouping operations. Familiar pandas workflows applied to biological data can yield new insights.